Spatial single-cell proteomics landscape decodes the tumor microenvironmental ecosystem of intrahepatic cholangiocarcinoma
Abstract
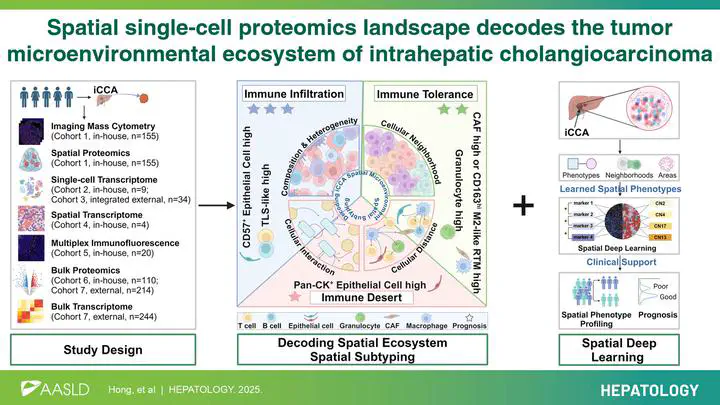
The prognoses and therapeutic responses of patients with intrahepatic cholangiocarcinoma (iCCA) are influenced by spatial interactions within the tumor microenvironment (TME), yet its spatial characteristics remain understudied. This study aimed to construct a comprehensive spatial atlas of iCCA using artificial intelligence-assisted spatial multiomics approaches and identify spatial features linked to prognosis and immunotherapy. By integrating data from diverse spatial multiomics technologies—including imaging mass cytometry, spatial proteomics, spatial transcriptomics, multiplex immunofluorescence, single-cell and bulk RNA/proteomics—across over 1.06 million resolved cells, we revealed that spatial topology, encompassing cellular deposition patterns, communities, and intercellular communications, strongly correlates with patient prognosis. Notably, CD163hi M2-like resident-tissue macrophages suppress antitumor immunity through direct interaction with CD8+ T cells, associating with poorer survival. We identified five spatial subtypes with distinct prognoses and proposed subtype-specific therapeutic strategies. Additionally, a spatial TME deep learning system was developed to accurately predict patient prognosis from a single 1-mm² tumor sample. This work provides foundational insights into the spatial TME ecosystem of iCCA, laying groundwork for precise patient stratification and personalized treatment development.