Leveraging Large Language Models for Early Diagnosis of Inherited Metabolic Diseases Evaluation and Optimization
Abstract
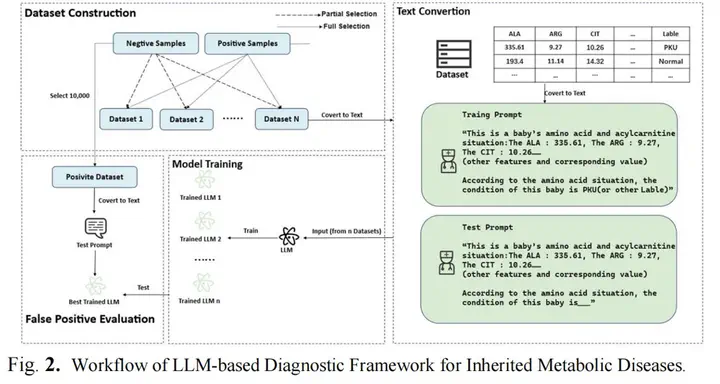
This study explores the feasibility of using large language models (LLMs) for early detection of neonatal genetic metabolic disorders, particularly their diagnostic efficiency for five amino acid metabolism-related diseases based on tandem mass spectrometry (MS/MS) data. By analyzing screening results from approximately 3,000 newborns, including 114 confirmed cases, the diagnostic accuracy of five LLMs, such as Qwen2.5-7B and Gemma-7B, was evaluated and compared with traditional machine learning methods. Fine-tuning was performed using quantization and Low-Rank Adaptation (LoRA) techniques to optimize key parameters such as the proportion of negative samples and the number of training iterations.Experimental results showed that LLMs outperformed traditional models in recall rates. Furthermore, the effect of quantization on model performance varied, with certain architectures maintaining high accuracy while significantly reducing computational overhead. False positive analysis revealed that LLMs effectively constrained the misclassification rate of healthy samples to below 2%, enhancing their practical value for large-scale screening. The study emphasizes the diagnostic robustness of LLMs in neonatal metabolic disorder screening and suggests that integrating multimodal patient data with clinical decision-support systems could further improve their applicability. However, issues such as data imbalance, computational costs, and model interpretability remain critical challenges that require further investigation